Apply any R function on rolling windows
Dawid Kałędkowski
2026-01-31
Source:vignettes/apply_any_r_function.Rmd
apply_any_r_function.RmdUsing runner
runner package provides functions applied on running
windows. The most universal function is runner::runner
which gives user possibility to apply any R function f on
running windows. Running windows are defined for each data window size
k, lag with respect to their indexes. Unlike
other available R packages, runner supports any input and
output type and also gives full control to manipulate window size and
lag/lead.
There are different kinds of running windows and all of them are
implemented in runner.
Cumulative windows
The simplest window type which is similar to
base::cumsum. At each element window is defined by all
elements appearing before current.
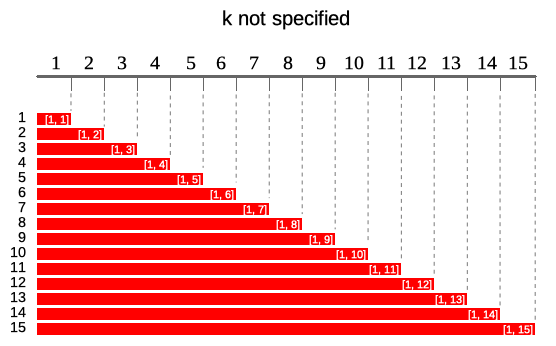
In runner this can be achieved as simple by:
Constant sliding windows
Second type of windows are these commonly known as
running/rolling/moving/sliding windows. This types of windows moves
along the index instead of cumulating like a previous one.
Following diagram illustrates running windows of length
k = 4. Each of 15 windows contains 4 elements (except first
three).
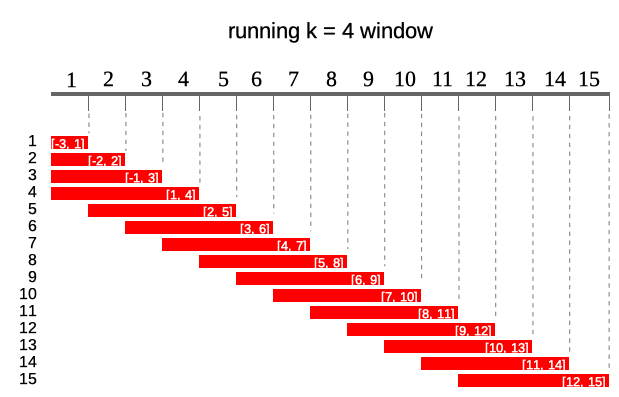
To obtain constant sliding windows one just needs to specify
k argument
# summarizing - sum of 4-elements
runner(
1:15,
k = 4,
f = sum
)
# summarizing - slope from lm
df <- data.frame(
a = 1:15,
b = 3 * 1:15 + rnorm(15)
)
runner(
x = df,
k = 5,
f = function(x) {
model <- lm(b ~ a, data = x)
coefficients(model)["a"]
}
)Windows depending on date
By default runner calculates on assumption that index
increments by one, but sometimes data points in dataset are not equally
spaced (missing weekends, holidays, other missings) and thus window size
should vary to keep expected time frame. If one specifies
idx argument, than running functions are applied on windows
depending on date rather on a sequence 1-n. idx should be
the same length as x and should be of type
Date, POSIXt or integer. Example
below illustrates window of size k = 5 lagged by
lag = 1. Note that one can specify also
k = "5 days" and lag = "day" as in
seq.POSIXt.
In the example below in square brackets ranges for each window.
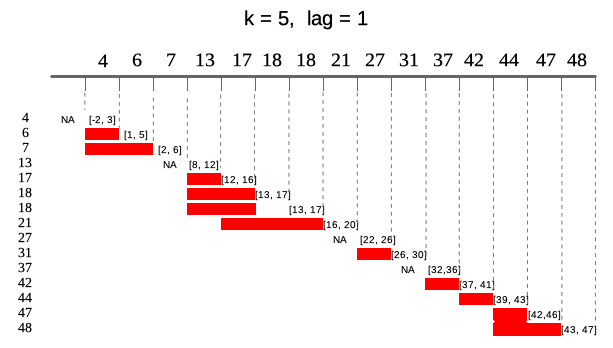
idx <- c(4, 6, 7, 13, 17, 18, 18, 21, 27, 31, 37, 42, 44, 47, 48)
# summarize - mean
runner::runner(
x = idx,
k = 5, # 5-days window
lag = 1,
idx = idx,
f = function(x) mean(x)
)
# use Date or datetime sequences
runner::runner(
x = idx,
k = "5 days", # 5-days window
lag = 1,
idx = Sys.Date() + idx,
f = function(x) mean(x)
)
# obtain window from above illustration
runner::runner(
x = idx,
k = "5 days",
lag = 1,
idx = Sys.Date() + idx
)running at
Runner by default returns vector of the same size as x
unless one puts any-size vector to at argument. Each
element of at is an index on which runner calculates
function. Example below illustrates output of runner for
at = c(13, 27, 45, 31) which gives windows in ranges
enclosed in square brackets. Range for at = 27 is
[22, 26] which is not available in current indices.
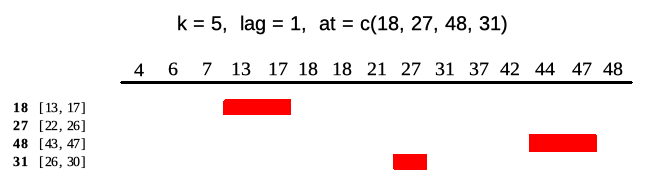
idx <- c(4, 6, 7, 13, 17, 18, 18, 21, 27, 31, 37, 42, 44, 47, 48)
# summary
runner::runner(
x = 1:15,
k = 5,
lag = 1,
idx = idx,
at = c(18, 27, 48, 31),
f = mean
)
# full window
runner::runner(
x = idx,
k = 5,
lag = 1,
idx = idx,
at = c(18, 27, 48, 31)
)at can also be specified as interval of the output
defined by time interval which results in obtaining results on following
indices
seq(min(idx), max(idx), by = "<time interval>").
Interval can be set in the same way as in seq.POSIXt
function. It’s worth noting that at interval shouldn’t be
more frequent than interval of idx - for Date
the most frequent interval is a "day", for
POSIXt it’s a "sec".
idx_date <- seq(Sys.Date(), Sys.Date() + 365, by = "1 month")
# change interval to 4-months
runner(
x = 0:12,
idx = idx_date,
at = "4 months"
)
# calculate correlation at every 6-months
runner(
x = data.frame(
a = 1:13,
b = 1:13 + rnorm(13, sd = 5),
idx_date
),
idx = "idx_date",
at = "6 months",
f = function(x) {
cor(x$a, x$b)
}
)Move and stretch window in time
One can stretch window length by k and shift in time (or
index) using lag. Both arguments can be
integer and also time interval like for example
2 months. If k or lag are a
single value then window size/lag are constant for all elements of x.
User can also specify k/lag as vector, then size and lag
will vary for each window. Both k and lag can
be of length(.) == 1, length(.) == length(x)
or length(.) == length(at) (if at is
specified). lag can be negative and positive while
k only non-negative.
# summarizing - concatenating
runner::runner(
x = 1:10,
lag = c(-1, 2, -1, -2, 0, 0, 5, -5, -2, -3),
k = c(0, 1, 1, 1, 1, 5, 5, 5, 5, 5),
f = paste,
collapse = ","
)
# full window
runner::runner(
x = 1:10,
lag = 1,
k = c(1, 1, 1, 1, 1, 5, 5, 5, 5, 5)
)
# on dates
idx <- c(4, 6, 7, 13, 17, 18, 18, 21, 27, 31, 37, 42, 44, 47, 48)
runner::runner(
x = 1:15,
lag = sample(c("-2 days", "-1 days", "1 days", "2 days"),
size = 15,
replace = TRUE
),
k = sample(c("5 days", "10 days", "15 days"),
size = 15,
replace = TRUE
),
idx = Sys.Date() + idx,
f = function(x) mean(x)
)
NA padding
Using runner one can also specify
na_pad = TRUE which would return NA for any
window which is partially out of range - meaning that there is no
sufficient number of observations to fill the window. By default
na_pad = FALSE, which means that incomplete windows are
calculated anyway. na_pad is applied on normal cumulative
windows and on windows depending on date. In example below two windows
exceed range given by idx so for these windows are empty
for na_pad = TRUE. If used sets na_pad = FALSE
first window will be empty (no single element within
[-2, 3]) and last window will return elements within
matching idx.

Using runner with data.frame
User can also put data.frame into x
argument and apply functions which involve multiple columns. In example
below we calculate beta parameter of lm model on 1, 2, …, n
observations respectively. On the plot one can observe how
lm parameter adapt with increasing number of
observation.
x <- cumsum(rnorm(40))
y <- 3 * x + rnorm(40)
date <- Sys.Date() + cumsum(sample(1:3, 40, replace = TRUE)) # unequaly spaced time series
group <- rep(c("a", "b"), 20)
df <- data.frame(date, group, y, x)
slope <- runner(
df,
function(x) {
coefficients(lm(y ~ x, data = x))[2]
}
)
plot(slope)One can also use runner with dplyr also
with problematic group_by operations, without need to apply
group_modify.
Below we apply grouped 20-days beta, by specifying window length
k = "10 days" and providing column name where indices
(dates) are kept.
library(dplyr)
summ <- df %>%
group_by(group) %>%
mutate(
cumulative_mse = runner(
x = .,
k = "20 days",
idx = "date", # specify column name instead df$date
f = function(x) {
coefficients(lm(y ~ x, data = x))[2]
}
)
)
library(ggplot2)
summ %>%
ggplot(aes(x = date, y = cumulative_mse, group = group, color = group)) +
geom_line()When user executes multiple runner calls in
dplyr mutate, one can also use run_by function
to prespecify arguments in tidyverse pipeline. In the
example below runner functions are applied on
k = "20 days" calculated on "date" column.
df %>%
group_by(group) %>%
run_by(idx = "date", k = "20 days", na_pad = FALSE) %>%
mutate(
cumulative_mse = runner(
x = .,
f = function(x) {
mean((residuals(lm(y ~ x, data = x)))^2)
}
),
intercept = runner(
x = .,
f = function(x) {
coefficients(lm(y ~ x, data = x))[1]
}
),
slope = runner(
x = .,
f = function(x) {
coefficients(lm(y ~ x, data = x))[2]
}
)
)Parallel mode
The runner function can also compute windows in parallel
mode. The function doesn’t initialize the parallel cluster automatically
but one have to do this outside and pass it to the runner
through cl argument.
library(parallel)
numCores <- detectCores()
cl <- makeForkCluster(numCores)
runner(
x = df,
k = 10,
idx = "date",
f = function(x) sum(x$x),
cl = cl
)
stopCluster(cl)Executing runner in parallel mode isn’t always
faster than a single thread. Multiple-thread computation
generates some overhead due to managing the nodes. In general,
complex functions which bases on processor (e.g. loops) used to be
quicker in parallel mode but one should assess itself which option
has the edge in specific situation.